paramz package¶
Subpackages¶
- paramz.core package
- Submodules
- paramz.core.constrainable module
- paramz.core.gradcheckable module
- paramz.core.index_operations module
- paramz.core.indexable module
- paramz.core.lists_and_dicts module
- paramz.core.nameable module
- paramz.core.observable module
- paramz.core.observable_array module
- paramz.core.parameter_core module
- paramz.core.parentable module
- paramz.core.pickleable module
- paramz.core.updateable module
- Module contents
- paramz.examples package
- paramz.optimization package
- paramz.tests package
- Submodules
- paramz.tests.array_core_tests module
- paramz.tests.cacher_tests module
- paramz.tests.examples_tests module
- paramz.tests.index_operations_tests module
- paramz.tests.init_tests module
- paramz.tests.lists_and_dicts_tests module
- paramz.tests.model_tests module
- paramz.tests.observable_tests module
- paramz.tests.parameterized_tests module
- paramz.tests.pickle_tests module
- paramz.tests.verbose_optimize_tests module
- Module contents
Submodules¶
paramz.__version__ module¶
paramz.caching module¶
-
class
Cache_this(limit=5, ignore_args=(), force_kwargs=())[source]¶ Bases:
objectA decorator which can be applied to bound methods in order to cache them
-
class
Cacher(operation, limit=3, ignore_args=(), force_kwargs=(), cacher_enabled=True)[source]¶ Bases:
objectCache an operation. If the operation is a bound method we will create a cache (FunctionCache) on that object in order to keep track of the caches on instances.
- Warning: If the instance already had a Cacher for the operation
- that Cacher will be overwritten by this Cacher!
Parameters: - operation (callable) – function to cache
- limit (int) – depth of cacher
- ignore_args ([int]) – list of indices, pointing at arguments to ignore in *args of operation(*args). This includes self, so make sure to ignore self, if it is not cachable and you do not want this to prevent caching!
- force_kwargs ([str]) – list of kwarg names (strings). If a kwarg with that name is given, the cacher will force recompute and wont cache anything.
- verbose (int) – verbosity level. 0: no print outs, 1: casual print outs, 2: debug level print outs
-
add_to_cache(cache_id, inputs, output)[source]¶ This adds cache_id to the cache, with inputs and output
-
combine_inputs(args, kw, ignore_args)[source]¶ Combines the args and kw in a unique way, such that ordering of kwargs does not lead to recompute
-
disable_cacher()[source]¶ Disable the caching of this cacher. This also removes previously cached results
-
on_cache_changed(direct, which=None)[source]¶ A callback funtion, which sets local flags when the elements of some cached inputs change
this function gets ‘hooked up’ to the inputs when we cache them, and upon their elements being changed we update here.
paramz.domains module¶
(Hyper-)Parameter domains defined for paramz.transformations.
These domains specify the legitimate realm of the parameters to live in.
_REAL:- real domain, all values in the real numbers are allowed
_POSITIVE:- positive domain, only positive real values are allowed
_NEGATIVE:- same as
_POSITIVE, but only negative values are allowed _BOUNDED:- only values within the bounded range are allowed, the bounds are specified withing the object with the bounded range
paramz.model module¶
-
class
Model(name)[source]¶ Bases:
paramz.parameterized.Parameterized-
objective_function()[source]¶ The objective function for the given algorithm.
This function is the true objective, which wants to be minimized. Note that all parameters are already set and in place, so you just need to return the objective function here.
For probabilistic models this is the negative log_likelihood (including the MAP prior), so we return it here. If your model is not probabilistic, just return your objective to minimize here!
-
objective_function_gradients()[source]¶ The gradients for the objective function for the given algorithm. The gradients are w.r.t. the negative objective function, as this framework works with negative log-likelihoods as a default.
You can find the gradient for the parameters in self.gradient at all times. This is the place, where gradients get stored for parameters.
This function is the true objective, which wants to be minimized. Note that all parameters are already set and in place, so you just need to return the gradient here.
For probabilistic models this is the gradient of the negative log_likelihood (including the MAP prior), so we return it here. If your model is not probabilistic, just return your negative gradient here!
-
optimize(optimizer=None, start=None, messages=False, max_iters=1000, ipython_notebook=True, clear_after_finish=False, **kwargs)[source]¶ Optimize the model using self.log_likelihood and self.log_likelihood_gradient, as well as self.priors.
kwargs are passed to the optimizer. They can be:
Parameters: - max_iters (int) – maximum number of function evaluations
- optimizer (string) – which optimizer to use (defaults to self.preferred optimizer)
Messages: True: Display messages during optimisation, “ipython_notebook”:
- Valid optimizers are:
- ‘scg’: scaled conjugate gradient method, recommended for stability.
- See also GPy.inference.optimization.scg
- ‘fmin_tnc’: truncated Newton method (see scipy.optimize.fmin_tnc)
- ‘simplex’: the Nelder-Mead simplex method (see scipy.optimize.fmin),
- ‘lbfgsb’: the l-bfgs-b method (see scipy.optimize.fmin_l_bfgs_b),
- ‘lbfgs’: the bfgs method (see scipy.optimize.fmin_bfgs),
- ‘sgd’: stochastic gradient decsent (see scipy.optimize.sgd). For experts only!
-
optimize_restarts(num_restarts=10, robust=False, verbose=True, parallel=False, num_processes=None, **kwargs)[source]¶ Perform random restarts of the model, and set the model to the best seen solution.
If the robust flag is set, exceptions raised during optimizations will be handled silently. If _all_ runs fail, the model is reset to the existing parameter values.
**kwargs are passed to the optimizer.
Parameters: - num_restarts (int) – number of restarts to use (default 10)
- robust (bool) – whether to handle exceptions silently or not (default False)
- parallel (bool) – whether to run each restart as a separate process. It relies on the multiprocessing module.
- num_processes – number of workers in the multiprocessing pool
- max_f_eval (int) – maximum number of function evaluations
- max_iters (int) – maximum number of iterations
- messages (bool) – whether to display during optimisation
Note
If num_processes is None, the number of workes in the multiprocessing pool is automatically set to the number of processors on the current machine.
-
paramz.param module¶
-
class
Param(name, input_array, default_constraint=None, *a, **kw)[source]¶ Bases:
paramz.core.parameter_core.Parameterizable,paramz.core.observable_array.ObsArParameter object for GPy models.
Parameters: - name (str) – name of the parameter to be printed
- input_array (np.ndarray) – array which this parameter handles
- default_constraint – The default constraint for this parameter
You can add/remove constraints by calling constrain on the parameter itself, e.g:
- self[:,1].constrain_positive()
- self[0].tie_to(other)
- self.untie()
- self[:3,:].unconstrain()
- self[1].fix()
Fixing parameters will fix them to the value they are right now. If you change the fixed value, it will be fixed to the new value!
Important Notes:
The array given into this, will be used as the Param object. That is, the memory of the numpy array given will be the memory of this object. If you want to make a new Param object you need to copy the input array!
Multilevel indexing (e.g. self[:2][1:]) is not supported and might lead to unexpected behaviour. Try to index in one go, using boolean indexing or the numpy builtin np.index function.
See
GPy.core.parameterized.Parameterizedfor more details on constraining etc.-
build_pydot(G)[source]¶ Build a pydot representation of this model. This needs pydot installed.
Example Usage:
np.random.seed(1000) X = np.random.normal(0,1,(20,2)) beta = np.random.uniform(0,1,(2,1)) Y = X.dot(beta) m = RidgeRegression(X, Y) G = m.build_pydot() G.write_png(‘example_hierarchy_layout.png’)
The output looks like:
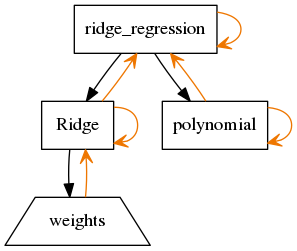
Rectangles are parameterized objects (nodes or leafs of hierarchy).
Trapezoids are param objects, which represent the arrays for parameters.
Black arrows show parameter hierarchical dependence. The arrow points from parents towards children.
Orange arrows show the observer pattern. Self references (here) are the references to the call to parameters changed and references upwards are the references to tell the parents they need to update.
-
copy()[source]¶ Returns a (deep) copy of the current parameter handle.
All connections to parents of the copy will be cut.
Parameters: - memo (dict) – memo for deepcopy
- which (Parameterized) – parameterized object which started the copy process [default: self]
-
parameter_names(add_self=False, adjust_for_printing=False, recursive=True, **kw)[source]¶ Get the names of all parameters of this model or parameter. It starts from the parameterized object you are calling this method on.
- Note: This does not unravel multidimensional parameters,
- use parameter_names_flat to unravel parameters!
Parameters: - add_self (bool) – whether to add the own name in front of names
- adjust_for_printing (bool) – whether to call adjust_name_for_printing on names
- recursive (bool) – whether to traverse through hierarchy and append leaf node names
- intermediate (bool) – whether to add intermediate names, that is parameterized objects
-
flattened_parameters¶
-
gradient¶ Return a view on the gradient, which is in the same shape as this parameter is. Note: this is not the real gradient array, it is just a view on it.
To work on the real gradient array use: self.full_gradient
-
is_fixed¶
-
num_params¶
-
param_array¶ As we are a leaf, this just returns self
-
parameters= []¶
-
values¶ Return self as numpy array view
-
class
ParamConcatenation(params)[source]¶ Bases:
objectParameter concatenation for convenience of printing regular expression matched arrays you can index this concatenation as if it was the flattened concatenation of all the parameters it contains, same for setting parameters (Broadcasting enabled).
See
GPy.core.parameter.Paramfor more details on constraining.-
constrain(constraint, warning=True)[source]¶ Parameters: - transform – the
paramz.transformations.Transformationto constrain the this parameter to. - warning – print a warning if re-constraining parameters.
Constrain the parameter to the given
paramz.transformations.Transformation.- transform – the
-
constrain_bounded(lower, upper, warning=True)[source]¶ Parameters: - upper (lower,) – the limits to bound this parameter to
- warning – print a warning if re-constraining parameters.
Constrain this parameter to lie within the given range.
-
constrain_fixed(value=None, warning=True, trigger_parent=True)[source]¶ Constrain this parameter to be fixed to the current value it carries.
This does not override the previous constraints, so unfixing will restore the constraint set before fixing.
Parameters: warning – print a warning for overwriting constraints.
-
constrain_negative(warning=True)[source]¶ Parameters: warning – print a warning if re-constraining parameters. Constrain this parameter to the default negative constraint.
-
constrain_positive(warning=True)[source]¶ Parameters: warning – print a warning if re-constraining parameters. Constrain this parameter to the default positive constraint.
-
fix(value=None, warning=True, trigger_parent=True)¶ Constrain this parameter to be fixed to the current value it carries.
This does not override the previous constraints, so unfixing will restore the constraint set before fixing.
Parameters: warning – print a warning for overwriting constraints.
-
unconstrain(*constraints)[source]¶ Parameters: transforms – The transformations to unconstrain from. remove all
paramz.transformations.Transformationtransformats of this parameter object.
-
unconstrain_bounded(lower, upper)[source]¶ Parameters: upper (lower,) – the limits to unbound this parameter from Remove (lower, upper) bounded constrain from this parameter/
-
unconstrain_fixed()[source]¶ This parameter will no longer be fixed.
If there was a constraint on this parameter when fixing it, it will be constraint with that previous constraint.
-
unfix()¶ This parameter will no longer be fixed.
If there was a constraint on this parameter when fixing it, it will be constraint with that previous constraint.
-
paramz.parameterized module¶
-
class
Parameterized(name=None, parameters=[])[source]¶ Bases:
paramz.core.parameter_core.ParameterizableSay m is a handle to a parameterized class.
Printing parameters:
- print m: prints a nice summary over all parameters - print m.name: prints details for param with name 'name' - print m[regexp]: prints details for all the parameters which match (!) regexp - print m['']: prints details for all parametersFields:
Name: The name of the param, can be renamed! Value: Shape or value, if one-valued Constrain: constraint of the param, curly "{c}" brackets indicate some parameters are constrained by c. See detailed print to get exact constraints. Tied_to: which paramter it is tied to.Getting and setting parameters:
- Set all values in param to one: m.name.to.param = 1 - Set all values in parameterized: m.name[:] = 1 - Set values to random values: m[:] = np.random.norm(m.size)
Handling of constraining, fixing and tieing parameters:
- You can constrain parameters by calling the constrain on the param itself, e.g: - m.name[:,1].constrain_positive() - m.name[0].tie_to(m.name[1]) - Fixing parameters will fix them to the value they are right now. If you change the parameters value, the param will be fixed to the new value! - If you want to operate on all parameters use m[''] to wildcard select all paramters and concatenate them. Printing m[''] will result in printing of all parameters in detail.
-
build_pydot(G=None)[source]¶ Build a pydot representation of this model. This needs pydot installed.
Example Usage:
np.random.seed(1000) X = np.random.normal(0,1,(20,2)) beta = np.random.uniform(0,1,(2,1)) Y = X.dot(beta) m = RidgeRegression(X, Y) G = m.build_pydot() G.write_png('example_hierarchy_layout.png')
The output looks like:
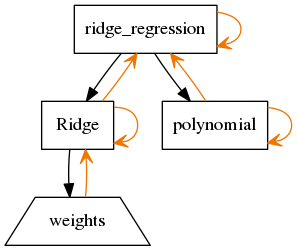
Rectangles are parameterized objects (nodes or leafs of hierarchy).
Trapezoids are param objects, which represent the arrays for parameters.
Black arrows show parameter hierarchical dependence. The arrow points from parents towards children.
Orange arrows show the observer pattern. Self references (here) are the references to the call to parameters changed and references upwards are the references to tell the parents they need to update.
-
copy(memo=None)[source]¶ Returns a (deep) copy of the current parameter handle.
All connections to parents of the copy will be cut.
Parameters: - memo (dict) – memo for deepcopy
- which (Parameterized) – parameterized object which started the copy process [default: self]
-
link_parameter(param, index=None)[source]¶ Parameters: - parameters (list of or one
paramz.param.Param) – the parameters to add - [index] – index of where to put parameters
Add all parameters to this param class, you can insert parameters at any given index using the
list.insertsyntax- parameters (list of or one
-
link_parameters(*parameters)[source]¶ convenience method for adding several parameters without gradient specification
-
unlink_parameter(param)[source]¶ Parameters: param – param object to remove from being a parameter of this parameterized object.
-
flattened_parameters¶
-
paramz.transformations module¶
-
class
Exponent[source]¶ Bases:
paramz.transformations.Transformation-
gradfactor(f, df)[source]¶ df(opt_param)_dopt_param evaluated at self.f(opt_param)=model_param, times the gradient dL_dmodel_param,
i.e.: define
\[\]rac{ rac{partial L}{partial f}left(left.partial f(x)}{partial x} ight|_{x=f^{-1}(f) ight)}
-
log_jacobian(model_param)[source]¶ compute the log of the jacobian of f, evaluated at f(x)= model_param
-
log_jacobian_grad(model_param)[source]¶ compute the drivative of the log of the jacobian of f, evaluated at f(x)= model_param
-
domain= 'positive'¶
-
-
class
Logexp[source]¶ Bases:
paramz.transformations.Transformation-
gradfactor(f, df)[source]¶ df(opt_param)_dopt_param evaluated at self.f(opt_param)=model_param, times the gradient dL_dmodel_param,
i.e.: define
\[\]rac{ rac{partial L}{partial f}left(left.partial f(x)}{partial x} ight|_{x=f^{-1}(f) ight)}
-
log_jacobian(model_param)[source]¶ compute the log of the jacobian of f, evaluated at f(x)= model_param
-
log_jacobian_grad(model_param)[source]¶ compute the drivative of the log of the jacobian of f, evaluated at f(x)= model_param
-
domain= 'positive'¶
-
-
class
Logistic(lower, upper)[source]¶ Bases:
paramz.transformations.Transformation-
gradfactor(f, df)[source]¶ df(opt_param)_dopt_param evaluated at self.f(opt_param)=model_param, times the gradient dL_dmodel_param,
i.e.: define
\[\]rac{ rac{partial L}{partial f}left(left.partial f(x)}{partial x} ight|_{x=f^{-1}(f) ight)}
-
domain= 'bounded'¶
-
-
class
NegativeExponent[source]¶ Bases:
paramz.transformations.Exponent-
gradfactor(f, df)[source]¶ df(opt_param)_dopt_param evaluated at self.f(opt_param)=model_param, times the gradient dL_dmodel_param,
i.e.: define
\[\]rac{ rac{partial L}{partial f}left(left.partial f(x)}{partial x} ight|_{x=f^{-1}(f) ight)}
-
domain= 'negative'¶
-
-
class
NegativeLogexp[source]¶ Bases:
paramz.transformations.Transformation-
gradfactor(f, df)[source]¶ df(opt_param)_dopt_param evaluated at self.f(opt_param)=model_param, times the gradient dL_dmodel_param,
i.e.: define
\[\]rac{ rac{partial L}{partial f}left(left.partial f(x)}{partial x} ight|_{x=f^{-1}(f) ight)}
-
domain= 'negative'¶
-
logexp= Logexp¶
-
-
class
Square[source]¶ Bases:
paramz.transformations.Transformation-
gradfactor(f, df)[source]¶ df(opt_param)_dopt_param evaluated at self.f(opt_param)=model_param, times the gradient dL_dmodel_param,
i.e.: define
\[\]rac{ rac{partial L}{partial f}left(left.partial f(x)}{partial x} ight|_{x=f^{-1}(f) ight)}
-
domain= 'positive'¶
-
-
class
Transformation[source]¶ Bases:
object-
gradfactor(model_param, dL_dmodel_param)[source]¶ df(opt_param)_dopt_param evaluated at self.f(opt_param)=model_param, times the gradient dL_dmodel_param,
i.e.: define
\[\]rac{ rac{partial L}{partial f}left(left.partial f(x)}{partial x} ight|_{x=f^{-1}(f) ight)}
-
log_jacobian(model_param)[source]¶ compute the log of the jacobian of f, evaluated at f(x)= model_param
-
log_jacobian_grad(model_param)[source]¶ compute the drivative of the log of the jacobian of f, evaluated at f(x)= model_param
-
domain= None¶
-